S.M. Hosseini-Mazinani,1 J. Koochmeshgi,1,2 Z. Khazaee-Koohpar,1 N. Hosein- Pur-Nobari1,2 and S.M. Seifati 1,2
كشف الحاملين لخلِّة بيلة الفنيل كيتون في الأسر الإيرانية بتحليل تعدد أشكال المتكررات المتـرادفة المتغيرة العدد
سيد مهدي حسيني مزيناني، جلال كوج مشكي، زينب خزابي كوهبر، نسرين حسين بور نوبري، سيد مرتضى سيفني
الخلاصـة: تـزداد وقوعات بيلة الفنيل كيتون، وهي إحدى الاضطرابات الصبغية الجسدية المتنحية، في إقليم شرق المتوسط، الذي يشيع فيه زواج الأقارب. وهذه الدراسة التي أجريت على الأسر الإيرانية، تقدِّم تقيـيماً لفائدة كشف الحاملين لخلة بيلة الفنيل كيتون، بتحليل تعدد أشكال المتكررات المتـرادفة المتغيرة العدد VNTR. وقام الباحثون بدراسة 171 شخصاً (45 منهم لا قرابةَ بينهم ولديهم بيلة فنيل كيتون مؤكدة كيميائياً حيوياً، وتم أيضاً فحص والديهم وأشقائهم غير المصابين). وتم التعرُّف على خمسة ألائل من المتكررات المتـرادفة ذات العدد المتغير، وذلك من بين 342 صبغياً (131 لا صلة لها ببيلة الفنيل كيتون و211 لها صلة بها). ومن شأن هذا النظام المتعلق بالمتكررات المتـرادفة ذات العدد المتغير أن يفضي إلى توفير محتوى معلوماتي حول تعدد الأشكال يصل إلى 66%، وهي نسبة تقارن بما لدى الأوربيين لكنها أكثر منها لدى الصينيين. وقد وفَّرت عملية كشف الحاملين من خلال التحليل المرتكز على فصل المتكررات المتـرادفة ذات العدد المتغير، معلومات جيدة في 89.5% من الأشقَّاء. ويستنتج الباحثون أن هذا التحليل الخاص بتعدد الأشكال يوفر معلومات أفضل كثيراً في عملية كشف حاملي بيلة الفنيل كيتون لدى السكان الإيرانيين.
ABSTRACT: This study of Iranian families assessed the usefulness of carrier detection of phenylketonuria by variable number tandem-repeat (VNTR) polymorphism analysis. We studied 171 people (45 unrelated PKU subjects, and their parents and unaffected siblings). Of 342 chromosomes (131 non-PKU and 211 PKU), 5 VNTR alleles were identified. This VNTR system would yield a polymorphism information content of 66%, comparable to that in Europeans and higher than in Chinese. Carrier detection by segregation analysis of VNTR was informative in 89.5% of siblings. We conclude that this polymorphism is highly informative in carrier detection of PKU in the Iranian population.
Détection des porteurs de phénylcétonurie dans des familles iraniennes par l’analyse du polymorphisme des séquences répétées en tandem en nombre variable
RÉSUMÉ: Cette étude sur des familles iraniennes a évalué l’utilité de la détection des porteurs de phénylcétonurie par l’analyse du polymorphisme des séquences répétées en tandem en nombre variable (variable number tandem repeats, ou VNTR). Nous avons étudié 171 personnes (45 sujets phénylcétonuriques sans lien de parenté ainsi que leurs parents et frères ou sœurs non atteints). Sur 342 chromosomes (131 non porteurs du gène muté responsable de la PCU et 211 porteurs de ce gène), 5 allèles VNTR ont été identifiés. Le système d’analyse des VNTR devrait produire un contenu informatif du polymorphisme de 66 %, chiffre comparable à celui observé chez les Européens et supérieur à celui observé chez les Chinois. La détection des porteurs par analyse de ségrégation des VNTR a permis d’obtenir des informations chez 89,5 % des frères et sœurs. Nous en concluons que ce polymorphisme apporte des informations très intéressantes aux fins de la détection des porteurs de PCU dans la population iranienne.
1National Research Centre for Genetic Engineering and Biotechnology, Tehran, Islamic Republic of Iran (Correspondence to S.M. Hosseini-Mazinani:
2Research Institute for Life, Tehran, Islamic Republic of Iran.
Received: 04/03/06; accepted: 03/07/06
EMHJ, 2008,14(6):1445-1451
Introduction
Phenylketonuria (PKU; McKusick 261600) was first described in 1934 by Følling [1,2]. It is caused by a deficiency of hepatic phenylalanine hydroxylase enzyme (PAH; EC 1.14.16.1). PAH is expressed in the liver and catalyzes the conversion of phenyl-alanine to tyrosine. The absence of PAH enzymatic activity causes persistent hyperphenylalaninaemia. In untreated individuals, PKU results in irreversible damage in postnatal brain development, which is clinically manifested as severe mental retardation [3].
PKU is an autosomal recessive disorder and the most prevalent disorder of amino acid metabolism. The overall incidence in Caucasians is estimated at 1 in 10 000 births, but is considerably higher in the Eastern Mediterranean region [3]. In fact, the highest incidence for PKU has been reported from this part of the world: 1 in 4000 in Turkey and 1 in 3627 in the Islamic Republic of Iran [4,5]. This high rate of incidence can be primarily attributed to the high degree of consanguineous marriage in the region [5,6].
Neonatal mass screening for PKU is a cost-effective and technically straightforward measure, which should be universally adopted in the region. Neonatal screening provides the opportunity for timely intervention in PKU. With the institution of a diet restricted in phenylalanine and supplemented by special protein substitutes, mental retardation can be prevented [3].
In view of the high degree of consanguinity in the Eastern Mediterranean region, carrier detection of PKU should also be considered as part of efforts to provide more accurate genetic counselling. The actual gene defect underlying PKU was elucidated in 1983 by Woo et al. [7]. Since then, several hundred mutations causing PKU have been characterized in the PAH gene [8,9]. Progress in assessing the usefulness of DNA analysis in the management of PKU has been obscured by numerous mutations of the PAH gene that have been described to date. Due to the large number of mutations, direct mutation analysis is not feasible in many cases, and indirect methods of segregation analysis of DNA polymorphisms within and in the vicinity of the gene assumes a major role in the carrier detection of PKU [10].
The PAH gene contains a multiallelic variable number tandem-repeat (VNTR) polymorphism, which is a 30 bp AT-rich tandem-repeat system located 3 kb down from the final exon of the gene [11]. This polymorphism is diversified enough in European Caucasians to be useful in carrier detection of PKU, with a polymorphism information content (PIC) of 70%. In Chinese PKU families, the PIC of this polymorphism is much lower, at 32% [10]. This polymorphism is also important in haplotyping the PAH gene and tracing the geographical origin of mutations [9,12,13].
To assess the usefulness of VNTR polymorphism in carrier detection of PKU in the Islamic Republic of Iran, we studied 45 PKU subjects and their parents and unaffected siblings, in accordance with established procedures of convenience sampling followed in other molecular genetic studies of PKU [e.g.: 14–16]. Results of these studies are usually taken to be indicative of the population under study. In view of the high degree of consanguinity in the Iranian population, care was taken to ensure that subjects were unrelated. The areas of residence of subjects represented a wide geographical distribution.
Methods
Sample
The study included 45 unrelated PKU subjects, ascertained biochemically, and their parents and unaffected siblings, a total of 171 people. The study was performed at the Department of Molecular Genetics, National Research Centre for Genetic Engineering and Biotechnology, Tehran, Islamic Republic of Iran, from 2000 to 2004. Subjects and their families had been referred in response to calls to PKU care centres throughout the country. Participation in the study was on a voluntary basis. Less than 20% of families referred declined to participate in the study and their decision was respected.
Data collection
Guidelines on ethical issues in medical genetics, as defined by the World Health Organization (WHO), were rigorously followed [17]. After explaining the study to participants and obtaining informed consent from them and their legal guardians, 3 mL of venous blood was taken from each participant. The carrier detection part of the study was restricted to competent adults and was performed under the supervision of qualified physicians and genetic counsellors, ensuring proper counselling and privacy of individuals.
PCR amplification
DNA fragments containing the PAH VNTR region were amplified by PCR in 20 μL of solution containing 25 ng genomic DNA, 10 pmol of each primer (VNTR-F: GCTTGAAACTTGAAAGTTGC; VNTR-R: GGAAACTTAAGAATCCCATC), 200 μM dNTP, 50 mM KCl, 10 mM tris–HCl (pH 8.4), and 1.5 mM MgCl2. Following an initial hot start (95 ºC, 5 min), Taq DNA polymerase was added (2 U, Cinagene Co. Tehran, Islamic Republic of Iran) and samples were subjected to 32 cycles of 1 min at 94 ºC, 1 min at 60 ºC, 1 min at 72 ºC, and a final elongation step (10 min at 72 ºC).
Gel electrophoresis
An aliquot of 5 mL of each polymerase chain reaction (PCR) product, along with a DNA size marker, were electrophoresed in separate lanes on 1.5% agarose gel. PCR products of 380, 470, 500, 530 and 560 base pairs corresponded to the presence of 3, 6, 7, 8 or 9 copies of the VNTR respectively. Routine analysis was performed on 12% polyacrylamide gel.
DNA sequencing
To verify the results, PCR products generated from samples homozygous for different VNTR genotypes were recovered from gel and sent for sequencing at the Center for Applied Genomics of the Hospital for Sick Children, Toronto, Canada.
Results
There were 45 unrelated PKU subjects and their parents and unaffected siblings in the study. Overall, 171 people were studied, with 342 chromosomes in total: 131 non-PKU chromosomes and 211 PKU chromosomes.
A total of 5 VNTR alleles were identified in Iranian PKU families, with VNTR repeats 3, 6, 7, 8 and 9 respectively. The distribution of alleles in non-PKU chromosomes was as follows: 45.0% VNTR3, 2.3% VNTR6, 12.2% VNTR7, 31.3% VNTR8 and 9.2% VNTR9. In PKU chromosomes, alleles were distributed as follows: 7.1% VNTR3, 0% VNTR6, 31.3% VNTR7, 48.3% VNTR8 and 13.3% VNTR9 (Table 1).
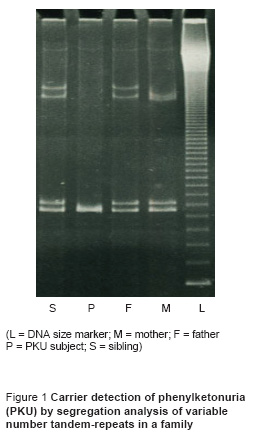
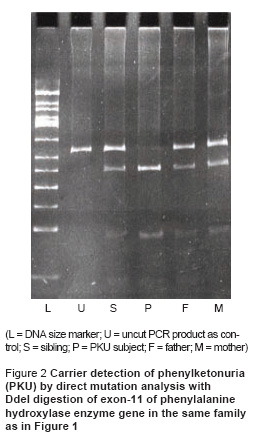
We performed carrier detection by segregation analysis of VNTR in 38 siblings of PKU subjects in 27 families. Wherever feasible, we confirmed the results by direct mutation analysis. The result of VNTR analysis was informative in 34 siblings (89.5%) and uninformative in 4 siblings (10.5%). VNTR analysis is illustrated for one family in Figure 1, indicating a carrier status in the sibling. Figure 2 shows the results of direct mutation analysis with DdeI digestion of exon-11 of the PAH enzyme gene in the same family as in Figure 1. The presence of this mutation creates a site of action for the DdeI restriction enzyme. The findings in Figure 1 are confirmed: both parents and sibling are heterozygous for the mutation.
Discussion
A total of 5 VNTR alleles were identified in Iranian PKU families: VNTR repeats 3, 6, 7, 8, and 9. The number of tandem repeats was confirmed by direct sequencing. The number of tandem repeats can also be deduced from the length of PCR products containing the VNTR region, as revealed by gel electrophoresis. In deducing the number of tandem repeats by gel electrophoresis, care must be taken to do this on agarose gel and not polyacrylamide gel. PCR products containing VNTR repeats tend to undergo conformation changes while running on polyacrylamide gel. This hampers their movement in the electrical field in comparison to the size marker. The fragments would appear larger than they really are and this can lead to spurious interpretation of results.
VNTR alleles identified in Iranian PKU families correspond to alleles previously described in European Caucasian families [10]. VNTR12, which comprises 4.0% of non-PKU alleles and 4.3% of PKU alleles in European Caucasians, was absent in our sample. On the other hand, diversity at this polymorphic locus was higher in our sample compared to Chinese PKU families, where only VNTR alleles 3 and 8 have been reported [10].
The frequencies of VNTR3 and VNTR7 in Iranian PKU chromosomes were markedly different from those reported for Europeans, with VNTR3 frequency being lower and that of VNTR7 higher than in European PKU chromosomes (Table 1). This can be partly attributed to different mutation spectra in these populations: R408W, the most prevalent PKU-causing mutation in European populations, is associated with VNTR3, particularly in the eastern parts of the continent [13]. This mutation is very rare in the Iranian population. On the other hand, IVS10nt546 “Mediterranean” mutation, associated with VNTR7, is the most prevalent mutation in the Islamic Republic of Iran (S.M. Hosseini-Mazinani, J. Koochmeshgi, unpublished data).
Assuming random mating, this VNTR system would yield a PIC of 66% in the Iranian population, comparable to that in Europeans (70%) and significantly higher than in the Chinese (32%) [10]. However, the high degree of consanguinity in this population must be taken into account, which results in strong deviation from the Hardy–Weinberg distribution.
Segregation analysis of VNTR in 38 siblings of PKU subjects in 27 families, confirmed by direct mutation analysis, was informative in 34 siblings (89.5%) and uninformative in 4 siblings (10.5%). Our results indicate that analysis of this polymorphism is highly informative in carrier detection of PKU in the Iranian population.
This VNTR polymorphism is part of a system of polymorphic markers comprising the haplotype of the PAH gene [9]. Other polymorphisms in this system include 7 restriction fragments length polymorphisms (RFLP) and a short tandem-repeat (STR) with a 4 bp repeat cassette. The VNTR polymorphism offers obvious advantages over these other polymorphisms in carrier detection of PKU. It is substantially more polymorphic than the dimorphic RFLPs, thereby increasing the likelihood of obtaining informative results in carrier detection studies. The STR site, like VNTR, is highly polymorphic. However, the small size of the STR repeat cassette makes it exceedingly difficult to differentiate alleles on routine gel electrophoresis studies, making STR a less attractive choice in carrier detection studies of PKU.
Conclusion
PKU is an exceedingly heterogeneous disorder in terms of its underlying mutations in the PAH gene. In many instances, the causative mutation is a rare one and cannot be readily identified. In these cases, the approach to carrier detection is based on segregation analysis of polymorphic markers associated with PAH. Our results indicate that the VNTR site of PAH gene is highly polymorphic in our population and its analysis can be considered the investigation of choice for carrier detection of PKU in instances where the cause of the PKU mutation is not known.
Acknowledgements
We wish to thank the Centre for Applied Genomics at the Hospital for Sick Children, Toronto, Canada, for their kind help in DNA sequencing.
Authornote
The first 2 listed authors contributed equally to this work.
References
- Christ SE. Asbjorn Følling and the discovery of phenylketonuria. Journal of the history of the neurosciences, 2003, 12(1):44–54.
- Følling A. Ueber Ausscheidung von Phenylbrenztraubensaeure in den Harn als Stoffwechselanomalie in Verbindung mit Imbezillitaet [The excretion of phenylpyruvic acid in the urine, an anomaly of metabolism in connection with imbecility]. Zeitschrift für physiologische Chemie, 1934, 227:169–76.
- Scriver CR, Kaufman S. Hyperphenylalaninemia: phenylalanine hydroxylase deficiency. In: Scriver CR et al., eds. The metabolic and molecular bases of inherited disease, 8th ed. New York, McGraw-Hill, 2001:1667–724.
- Ozalp I et al. Inherited metabolic disorders in Turkey. Journal of inherited metabolic disease, 1990, 13(5):732–8.
- Koochmeshgi J, Bagheri A, Hosseini-Mazinani SM. Incidence of phenylketonuria in Iran estimated from consanguineous marriages. Journal of inherited metabolic disease, 2002, 25(1):80–1.
- Woolf LI. Phenylketonuria in Turkey, Ireland, and West Scotland. Journal of inherited metabolic disease, 1994, 17(2):246–7.
- Woo SL et al. Cloned human phenylalanine hydroxylase gene allows prenatal diagnosis and carrier detection of classical phenylketonuria. Nature, 1983, 306(5939):151–5.
- Scriver CR et al. PAHdb 2003: what a locus-specific knowledgebase can do. Human mutation, 2003, 21(4):333–44.
- Phenylalanine hydroxylase locus knowledgebase [website] (http://www.pahdb.mcgill.ca/, accessed 17 January 2007).
- Eisensmith CR, Goltsov AA, Woo SLC. A simple, rapid, and highly informative PCR-based procedure for prenatal diagnosis and carrier screening of phenylketonuria. Prenatal diagnosis, 1994, 14(12):1113–8.
- Goltsov AA et al. Association between mutations and a VNTR in the human phenylalanine hydroxylase gene. American journal of human genetics, 1992, 51(3):627–36.
- Cali F et al. The STR252-IVS10nt546-VNTR7 phenylalanine hydroxylase minihaplotype in five Mediterranean samples. Human genetics, 1997, 100(3–4):350–5.
- Tighe O et al. Genetic diversity within the R408W phenylketonuria mutation lineages in Europe. Human mutation, 2003 21(4):387–93.
- Ozguc M et al. Mutation analysis in Turkish phenylketonuria patients. Journal of medical genetics, 1993, 30(2):129–30.
- Effat L et al. Haplotypes and mutations of the PAH locus in Egyptian families with PKU. European journal of human genetics, 1999, 7(2):259–62.
- Zschocke J et al. The molecular basis of phenylalanine hydroxylase deficiency in Croatia. Human mutation, 2003, 21(4):399.
- Wertz DC, Fletcher JC, Berg K, eds. Review of ethical issues in medical genetics. Geneva, World Health Organization, 2003 (WHO/HGN/ETH/00.4).








