O. Zouhairi,1 I. Saleh,1 N. Alwan,1 I. Toufeili,2 E. Barbour 3 and S. Harakeh1
المقاومة لمضادات الجراثيم في أنواع العنقوديات المستفردة من منتجات الألبان اللبنانية
عمر زهيري، إيمان صالح، نسرين علوان، عماد طفيلي، إيلي بربور، ستيف حركه
الخلاصـة: يقيِّم الباحثون في هذه الدراسة المقاومة لمضادات الجراثيم لدى ذراري ذات مواصفات جزيئية معروفة من العنقوديات الذهبية والعنقوديات الرِّمِّية المستفردة من ثلاثة منتجات طعامية لبنانية من الألبان التي قد تستهلك نيئة، وهي الكشك، والشنكليش، والجبن البلدي. وكان الباحثون يتعرّفون على المستفرَدات المشتبهة بكونها من العنقوديات بشكل مبدئي باستخدام اختبارات كيميائية حيوية معيارية، ثم يقيِّمون الذراي التي يؤكِّد التفاعل السلسلي للبوليميراز (وهي 29 من ذراري العنقودية الذهبية و17 من ذراري العنقودية الرمية) من حيث حساسيّتها لمختلف مضادات المكروبات. وتبيَّن الباحثون أن أعلى مستويات التلوث بالعنقوديات كانت في الجبن البلدي. وقد تراوحت معدّلات المقاومة بين %67 تجاه الجنتاميسين و%94 تجاه الأوكساسيلين والكلينداميسين. وتدلّ النتائج على أن الأغذية المنتَجَة محلياً من الألبان قد تؤدِّي دور الناقل لأنواع العنقوديات المقاومة لمضادات الجراثيم.
ABSTRACT The study evaluated the antimicrobial resistance of molecularly characterized strains of Staphylococcus aureus and S. saprophyticus isolated from 3 Lebanese dairy-based food products that are sometimes consumed raw: kishk, shanklish and baladi cheese. Suspected Staphylococcus isolates were identified initially using standard biochemical tests, then strains that were confirmed by polymerase chain reaction (29 S. aureus and 17 S. saprophyticus) were evaluated for their susceptibility to different antimicrobials. The highest levels of contamination with staphylococci were in baladi cheese. Resistance rates ranged from 67% to gentamicin to 94% to oxacillin and clindamycin. The results suggest that these locally made dairy-based foods may act as vehicles for the transmission of antimicrobial-resistant Staphylococcus spp.
Résistance aux antimicrobiens des espèces du genre Staphylococcus isolées dans des produits libanais dérivés du lait
RÉSUMÉ La présente étude a évalué la résistance aux antimicrobiens, après caractérisation moléculaire, de souches de Staphylococcus aureus et S. saprophyticus isolées à partir de trois produits libanais dérivés du lait, parfois consommés crus : le kishk, le shanklish et le fromage baladi. Des isolats suspects de Staphylococcus ont d’abord été identifiés par l’analyse biochimique classique, puis les souches confirmées par amplification en chaîne par polymérase (29 isolats de S. aureus et 17 isolats de S. saprophyticus) ont été analysées pour déterminer leur sensibilité aux différents antimicrobiens. Les taux de contamination par staphylocoques les plus élevés ont été retrouvés dans le fromage baladi. Les taux de résistance allaient de 67 % pour la gentamicine, à 94 % pour l’oxacilline et la clindamycine. Les résultats suggèrent que ces denrées dérivées du lait et produites localement pourraient être des vecteurs de la transmission des espèces Staphylococcus résistantes aux antimicrobiens.
1Department of Biology; 2Department of Nutrition and Food Science; 3Department of Animal and Veterinary Sciences, American University of Beirut, Beirut, Lebanon (Correspondence to S. Harakeh:
Received: 31/03/09; accepted: 03/06/09
EMHJ, 2010, 16(12): 1221-1225
Introduction
Milk and milk-based food products are highly susceptible to microbial attack because of their rich composition, which provides a favourable medium for growth of a host of spoilage agents [1]. Unpasteurized milk may become contaminated with enterotoxigenic coagulase-positive Staphylococcus spp. [2], either through contact with the cow’s udder during milking or by cross-contamination during processing [3]. Symptoms such as nausea, vomiting, abdominal cramps and diarrhoea usually appear 1–6 hours after ingestion of the enterotoxins produced in contaminated milk [4]. Consequently, enumeration and identification of staphylococci in dairy products is a priority in developing public health measures to reduce food-borne disease outbreaks [5,6].
The uncontrolled application of antimicrobials in the environment is leading to a constant increase in the rate of antimicrobial resistance among community-acquired staphylococci [7,8]. Staphylococcus spp. can rapidly acquire resistance to a broad range of antimicrobials, thereby posing a major concern in the treatment of staphylococcal infections [9]. Studying antimicrobial resistance in humans and animals is important for detecting changing patterns of resistance, implementing control measures on the use of antimicrobial agents and preventing the spread of multidrug-resistant strains of bacteria [10].
In Lebanon, Ministry of Public Health data showed an increase in the number of reported cases of food poisoning, from 43 in 2002 to 373 in 2004. Some of these cases were related to the consumption of homemade cheese [11]. The Lebanese Bekaa valley is known for its production of a wide range of dairy products that are consumed all over Lebanon. Most of these foods are produced in “cottage industry” conditions using traditional techniques with little emphasis on hygiene practices. The most common Lebanese dairy products are baladi cheese (white, semi-soft cheese), kishk (dried, fermented milk–wheat mixture) and shankleesh (cheese balls). The objective of this preliminary study was to assess the prevalence and antimicrobial resistance of S. aureus and S. saprophyticus in baladi cheese, kishk and shankleesh. Identification of these pathogens is important for surveillance, prevention and control of milk-borne diseases.
Methods
Baladi cheese, shankleesh and kishk were collected randomly from the Bekaa valley area of north-east Lebanon. Samples were collected on 4 trips between the months of August and December 2004. Target locations for sample collection included markets, houses and small family farms. In total, 164 samples were collected (83 kishk, 45 baladi cheese and 36 shankleesh). All samples were packaged in sterile bags and kept on ice in a refrigerator until brought to the laboratory. Samples were analysed within 24 hours.
Samples were diluted and macerated in a stomacher for 3 minutes. Further serial dilutions of samples were inoculated on duplicate plate count agar (for aerobic plate counts), McConkey agar or violet-red bile agar plates (Oxoid) (for total coliform counts) [12], and mannitol salt agar (for Staphylococcus spp.) (Oxoid) [7]. Plates were incubated at 37 °C for 24 hours.
Identification was carried out using standard methods [13]. All purple colonies on McConkey agar were considered as coliforms. Golden-yellow colonies surrounded by a yellow halo and white mucoid colonies were considered possible S. aureus and S. saprophyticus respectively, and were selected for further biochemical and molecular testing. Positive controls were performed by inoculating sterile milk with S. aureus, and S. saprophyticus obtained from the American type culture collection (ATCC), cultured, plated and incubated for 24 hours at 37 °C.
Based on Staphylococcus spp. counts, all samples were classified according to Public Health Laboratory Service guidelines [14] as satisfactory, acceptable, unsatisfactory and potentially hazardous. In the case of Staphylococcus spp., a sample was considered potentially hazardous if the bacterial count exceeded 103 colony forming units/g.
Suspected colonies were characterized biochemically. Presumptive staphylococcal colonies were confirmed by Gram staining. All Gram-positive cocci were divided into species on the basis of the type of haemolysis on blood agar. Colonies showing no haemolysis were suspected to be S. saprophyticus and subjected to PCR confirmation. Isolates showing α-haemolysis were further tested using a latex agglutination test (Pastorex Staph-Plus, BioRad), which is indicative of the presence of S. aureus [15]. PCR was used for further molecular confirmation of the positive isolates [16].
Extraction of DNA from both species of Gram-positive staphylococci was performed using the GFX Genomic Blood DNA Purification Kit (Amersham Biosciences). Amplification was performed from purified genomic DNA. Two sets of primers were used for the detection of Staphylococcus spp. For S. aureus detection, the nucA primer pair, designed by Palomares et al. was used to amplify a 279-bp fragment within the nucA gene. For S. saprophyticus, a primer pair yielding a 380-bp product was used [17]. Negative controls (no DNA template added) and positive controls (containing template DNA from S. aureus, or S. saprophyticus) were included in each PCR reaction performed. An aliquot of 10 μL of each PCR-amplified sample was mixed with 2 μL of 6× loading dye (BioRad) and loaded onto a 1.5% agarose gel stained with 0.25 μg ethidium bromide. Electrophoresis of the amplified DNA fragments was carried out in 0.04 M Tris-acetate/0.001 M EDTA at a constant voltage of 90 V for 100 minutes. The gel was visualized by UV illumination and photographed [8].
Positively identified Staphylococcus strains were tested for their susceptibility to different antimicrobials using the disk diffusion method with incubation at 37 °C overnight [18]. Six different antimicrobials were used: oxacillin (1 μg), teicoplanin (30 μg), gentamicin (10 μg), clindamycin (2 μg), vancomycin (30 μg) and methicillin (5 μg) (disks supplied by BioMerieux). Isolates were described either as resistant (not inhibited), or sensitive (appropriately inhibited) based on the size of the inhibition zone [18].
Statistical analysis
Statistical analyses were carried out using SPSS, version 11.0. The chi-squared test was used to compare the percentages of contamination of the 3 different dairy products tested.
Results
A total of 321 colonies were identified morphologically as suspected Staphylococcus spp. and were subjected to Gram staining; 254 were identified as Gram-positive cocci. Of these, 78 isolates showed no haemolysis on blood agar and were presumed to be S. saprophyticus. A total of 146 isolates showed complete lysis on blood agar, which is a typical of S. aureus. Of these, 41 isolates were positive by latex agglutination test and were presumed to be S. aureus. Isolates showing partial haemolysis on blood agar were discarded. Presumed S. saprophyticus and S. aureus isolates were kept and subjected to PCR using specific primers.
Of the 41 suspected S. aureus isolates, 29 were confirmed by PCR. All 29 isolates gave a clear 279 bp band, with the nucA primer pair (Figure 1). For S. saprophyticus, 17 of the 78 suspected isolates were confirmed using PCR. All 17 gave a 380 bp band with the sta-sap primer pair.
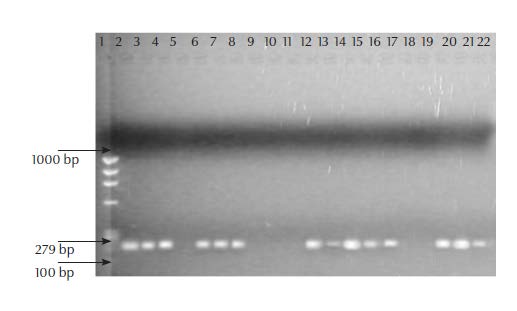
Of the 46 PCR-confirmed staphylococci isolates, 45 (97.8%) showed resistance to 1 or more antimicrobials while 61.0%, mostly S. aureus, showed resistance to all 6 antimicrobials. The isolates showed highest resistance to oxacillin (93.5%) and clindamycin (93.5%), followed by methicillin (84.8%), teicoplanin (76.1%), vancomycin (71.7%) and gentamicin (67.4%).
Microbiological quality of the tested dairy products
Bacterial counts
All the cheese, shankleesh and kishk samples had high aerobic plate counts; 76% of baladi cheese samples, 67% of shankleesh samples and 64% of kishk samples were contaminated with Staphylococcus spp. According to the guidelines provided by the Public Health Laboratory Service (PHLS), a sample is considered unfit for consumption if it contains only 1 colony of Staphylococcus spp. in 25g of sample. Therefore, all baladi cheese, shankleesh and kishk samples that showed the presence of Staphylococcus spp. were graded as potentially hazardous. Cheese samples were of significantly lower microbiological quality compared with the other 2 dairy products (P < 0.05).
PCR confirmation
Out of the 45 baladi cheese samples, 19 (42.2%) were positive for S. aureus and 3 (6.7%) were contaminated with S. saprophyticus based on PCR analysis. Only 1 baladi cheese sample was contaminated with both Staphylococcus spp. With respect to the 36 shankleesh samples, 2 (5.6%) tested positive for S. aureus and 9 (25.0%) for S. saprophyticus. Only 8 (9.6%) of the 83 kishk samples were contaminated with S. aureus, whereas S. saprophyticus was isolated from only 5 samples (6.0%).
Discussion
Traditional handmade dairy products may be produced from raw milk heated for only a few minutes to temperatures that are not enough to kill many pathogenic bacteria. The continuous increase in food poisoning cases reported around Lebanon has highlighted the need to monitor the manufacturing of food products in order to avoid future health hazards [11]. Milking operations, including storage, handling and transport of milk to the consumer or the factory, are considered critical points where contamination may occur. The use of mechanical milking equipment and improved sanitary practices has reduced the opportunity for contamination of milk by humans [19]. If raw milk is not rapidly refrigerated, then S. aureus can grow and produce a heat-stable enterotoxin that is usually not destroyed by pasteurization [20]. Staphylococci are halophilic bacteria and are able to survive in conditions of very low moisture content. This microorganism grows best on proteinaceous foods such as meat, poultry, milk and their products.
With respect to the 3 dairy food types studied, contamination could have occurred at any step of production. One of the crucial steps in the production of kishk and shankleesh is drying in the open air. At this point, microbial contamination is likely to occur. Production of kishk involves rubbing of the mix by hand which provides ample opportunity for bacterial contamination. The major sources of contamination in production of baladi cheese are the milk and manual handling at the different stages of processing.
The 3 tested dairy products showed variability in their microbiological qualities that may be attributed to differences in their chemical and physical compositions. Shankleesh is usually coated with thyme, which has been demonstrated to show inhibitory effects against bacteria [21]. Moreover, shankleesh is stored in olive oil, which makes the medium anaerobic thus inhibiting the growth of all aerobic pathogens [22]. The low moisture content of kishk (< 10%), its acidic nature (pH ~ 3.8) and its high salt level (~ 2.8 g NaCl/100 g of the dried product) prevent the growth of most bacterial species [23]. Cheese, on the other hand, is highly susceptible to microbial attack due to its high moisture content and moderate levels of salt.
Foodborne diseases, and especially milk-related infections, are not limited to poorer countries. Even in industrialized countries it has been reported that around 2%–6% of the bacterial outbreaks in which the food vehicle is known were related to milk and dairy products [1]. In Lebanon, no previous studies were conducted to evaluate the contamination level of dairy-based foods. Most of the previous studies in Arab countries have investigated clinical isolates. However, recent research conducted in Egypt on 152 samples of dairy-based products resulted in the identification of 16 Cronobacter strains, of which 3 showed resistance to trimethoprim and/or neomycin [24]. The results of that study together with our results show that dairy-based foods might act as sources of contamination for different bacterial strains. Therefore more studies and continuous evaluation of the microbiological quality of dairy products in the region are needed.
The emergence of antimicrobial-resistant bacteria is common to areas where antimicrobials are carelessly applied. One of the main reasons for the increase in environmental multi-drug resistant bacteria is the indiscriminate use of antimicrobials during animal husbandry. Because of the emergence of such bacteria, antimicrobials used in fighting infections are less effective. It is important to note that S. aureus can easily acquire resistance to most antimicrobials [9].
In Lebanon, the antimicrobial susceptibility of Gram-negative bacteria has been largely determined for clinical isolates [25]. However, very little is known about antimicrobial resistance patterns of environmental isolates. In our study, it was striking to note that 97.8% of the tested staphylococcal isolates were resistant to at least 1 antimicrobial. Resistance patterns ranged from 67.4% for gentamicin and 93.5% for both oxacillin and clindamycin. The high resistance towards oxacillin can be explained by the fact that resistance to oxacillin might be either chromosome- or plasmid-encoded; therefore, it can easily be acquired in nature by horizontal gene transfer [26]. The fact that resistance is high in environmental isolates is mainly because antimicrobials are frequently prescribed by veterinarians as treatment for Gram-negative bacterial infections on farms. Thus, the indiscriminate use of those antimicrobial agents might account, at least in part, for such a high resistance.
This study provides some important baseline data about the contamination status of dairy-based food products in Lebanon and the patterns of resistance of Staphylococcus spp. towards commonly used antimicrobials. The presence of multi-drug resistant strains is alarming, because such strains are considered a serious danger to public health. Additional research is required to better define the ecology and evolution of bacterial resistance to antimicrobial agents in the environment as a whole. Further research and subsequent management of this problem is vital to help ensure that the emergence of drug-resistant bacteria is limited and that antimicrobial agents remain effective.
References
- De Buyser ML et al. Implication of milk and milk products in food-borne diseases in France and in different industrialised countries. International Journal of Food Microbiology, 2001, 67:1–17.
- Simeão do Carmo L et al. Food poisoning due to enterotoxigenic strains of Staphylococcus present in Minas cheese and raw milk in Brazil. Food Microbiology, 2002, 19:9–14.
- Ramesh A et al. Application of a convenient DNA extraction method and multiplex PCR for the direct detection of Staphylococcus aureus and Yersinia enterocolitica in milk samples. Molecular and Cellular Probes, 2002, 16:307–314.
- Asao T et al. An extensive outbreak of staphylococcal food poisoning due to low-fat milk in Japan: estimation of enterotoxin A in the incriminated milk and powdered skim milk. Epidemiology and Infection, 2003, 130:33–40.
- Ercolini D et al. PCR-based detection of enterotoxigenic Staphylococcus aureus in the early stages of raw milk cheese making. Journal of Applied Microbiology, 2004, 96:1090–1096.
- Palomares C et al. Rapid detection and identification of Staphylococcus aureus from blood culture specimens using real-time fluorescence PCR. Diagnostic Microbiology and Infectious Disease, 2003, 45:183–189.
- Harakeh S, Yassine H, El-Fadel M. Antimicrobial-resistant patterns of Escherichia coli and Salmonella strains in the aquatic Lebanese environments. Environmental Pollution, 2006, 143:269–277.
- Harakeh S et al. Isolation, molecular characterization and antimicrobial resistance patterns of Salmonella and Escherichia coli isolates from meat-based fast food in Lebanon. Science of the Total Environment, 2005, 341:33–44.
- Bozdogan B et al. Derivatives of a vancomycin-resistant Staphylococcus aureus strain isolated at Hershey Medical Center. Antimicrobial Agents and Chemotherapy, 2004, 48:4762–4765.
- van Duijkeren E et al. Antimicrobial susceptibilities of Salmonella strains isolated from humans, cattle, pigs, and chickens in the Netherlands from 1984 to 2001. Journal of Clinical Microbiology, 2003, 41:3574–3578.
- Ministry of Public Health in Lebanon [website] (http://www.public-health.gov.lb, accessed 12 September 2010).
- Hayes MC et al. Identification and characterization of elevated microbial counts in bulk tank raw milk. Journal of Dairy Science, 2001, 84(1):292–298.
- Downes FP, Ito K, eds. Compendium of methods for the microbiological examination of foods, 4th ed. Washington, DC, American Public Health Association, 2002.
- Gilbert RJ et al. PHLS Advisory Committee for Food and Dairy Products. Guidelines for the microbiological quality of some ready-to-eat foods sampled at the point of sale. Communicable Disease and Public Health, 2000, 3:163–167.
- Rodrigues AG et al. Expression of plasma coagulase among pathogenic Candida species. Journal of Clinical Microbiology, 2003, 41:5792–5793.
- Cocolin L et al. Direct identification in food samples of Listeria spp. and Listeria monocytogenes by molecular methods. Applied and Environmental Microbiology, 2002, 68:6273–6282.
- Martineau F et al. Development of a rapid PCR assay specific for Staphylococcus saprophyticus and application to direct detection from urine samples. Journal of Clinical Microbiology, 2000, 38:3280–3284.
- Wikler MA. . Performance standards for antimicrobial disk susceptibility test. Approved standard, 10th ed. Wayne, Pennsyvania, Clinical and Laboratory Standards Institute, 2008.
- Kyozaire JK et al. Microbiological quality of goat’s milk obtained under different production systems. Journal of the South African Veterinary Association, 2005, 76:69–73.
- Bennett RW. Staphylococcal enterotoxin and its rapid identification in foods by enzyme-linked immunosorbent assay-based methodology. Journal of Food Protection, 2005, 68:1264–1270.
- Jugl-Chizzola M et al. Effects of Thymus vulgaris L. as feed additive in piglets and against haemolytic E. coli in vitro. Berliner und Munchener Tierarztliche Wochenschrift, 2005, 118:495–501.
- Toufeili I et al. Ripening changes and sensory properties of bovine, carpine, and ovine shankleesh. International Dairy Journal, 1995, 5:179–189.
- Tamime AY, McNulty D. Kishk—a dried fermented milk/cereal mixture. 4. Microbiological quality. Le Lait, 1999, 79:449–456.
- El-Sharoud WM et al. Characterization of Cronobacter recovered from dried milk and related products. BMC Microbiology, 2009, 9:24.
- Araj GF. Available laboratory tests to guide antimicrobial therapy. Lebanese Medical Journal, 2000, 48:199–202.
- Fluit AC, Verhoef J, Schmitz FJ. European SENTRY Participants. Frequency of isolation and antimicrobial resistance of Gram-negative and Gram-positive bacteria from patients in intensive care units of 25 European university hospitals participating in the European arm of the SENTRY Antimicrobial Surveillance Program 1997–1998. European Journal of Clinical Microbiology and Infectious Diseases, 2001, 20:617–625.








