A.A. El Kholy,1 H.E. Gomaa,2 M.G. El Anany1 and E. Abd El Rasheed2
تشخيص داء البروسيلات البشري في مصر بتفاعل سلسلة البوليميراز
أماني علي الخولي، هويدا عز الدين جمعة، مرفت جابر العناني، إيناس عبد الرشيد
الخلاصـة: تهدف هذه الدراسة لإيجاد تقنية لتفاعل سلسلة البوليميراز تتمتع بالحساسية وبالدقة لتشخيص داء البروسيلات البشري النشط في مصر. وقد فشل الباحثون في استخلاص دنا البروسيلات في عتيدة تجارية، إلا أنهم نجحوا في ذلك بواسطة عتيدة استخلاص صمموها في المختبر باستخدام مجموعتين من الـمَشْرَعات هما [(223bp) B5/B4 وJPR/JPF (193bp)]، وكان ذلك أكثر توفيراً من الناحية الاقتصادية. وقد أظهرت هذه التقنية درجة رفيعة من الحساسية والنوعية والدقة؛ فقد زادت إيجابية التفاعل زيادة ملحوظة مع زيادة عيارات الحساسية المصلية باستخدام اختبار التـراص في الأنابيب، وأظهر إيجابية %100 لدى المرضى الذين كان لديهم مزارع الدم إيجابية. ويوصي الباحثون باستخدام تفاعل سلسلة البوليميراز كبديل للزرع في تشخيص داء البروسيلات.
ABSTRACT This study aimed to establish an accurate and sensitive polymerase chain reaction (PCR) technique for the diagnosis of active human brucellosis in Egypt. We failed to extract Brucella DNA with a commercial kit, but an extraction kit designed in-house using 2 sets of primers [B4/B5 (223 bp) and JPF/JPR (193 bp)] was successful and more economical. The technique showed high sensitivity, specificity and accuracy. The PCR positivity increased significantly with increasing seropositivity titres by the standard tube agglutination test and showed 100% positivity in patients with positive blood cultures. We recommend using PCR as an alternative to culture for diagnosis of brucellosis.
Diagnostic de la brucellose humaine en Égypte au moyen de l’amplification en chaîne par polymérase
RÉSUMÉ, Cette étude visait à mettre en place une technique d’amplification en chaîne par polymérase (PCR) précise et sensible aux fins du diagnostic de la brucellose humaine évolutive en Égypte. Nous n’avons pas réussi à extraire l’ADN de Brucella avec un kit disponible dans le commerce, mais avec un kit d’extraction conçu en interne et utilisant deux jeux d’amorces [B4/B5 (223 pb) et JPF/JPR (193 pb)], qui s’est avéré efficace et plus économique. Cette technique a offert des niveaux de sensibilité, de spécificité et de précision élevés. La positivité de la PCR augmentait de façon significative avec l’augmentation des taux de séropositivité d’après le test classique d’agglutination en tube et s’élevait à 100 % chez les patients dont les hémocultures étaient positives. Nous recommandons d’utiliser la PCR comme solution autre que la culture pour le diagnostic de la brucellose.
1Department of Clinical and Chemical Pathology, Faculty of Medicine, University of Cairo, Cairo, Egypt.
2Department of Clinical and Chemical Pathology, National Research Centre, Cairo, Egypt (Correspondence to H.E. Gomaa:
Received: 28/11/06; accepted: 15/05/07
EMHJ, 2009, 15(5):1068-1074
Introduction
Brucellosis is an important health problem in Egypt and a confirmed cause of 3% of cases of acute febrile illness [1].
The high frequency of brucellosis and the nonspecific clinical picture emphasizes the importance of laboratory-based diagnosis. Blood culture is the gold standard for microbial diagnosis. However, it may take 2–6 weeks to isolate the organism. Also, prior use of antibiotics may interfere with growth of Brucella spp. Blood cultures require containment level 3 facilities to avoid laboratory-acquired infection, which may limit its use in small and remote health care facilities [2].
Several serological assays have been developed to diagnose brucellosis. Standard tube agglutination (STA)—the Wright test—is still the most reliable method [3]. Brucellosis can be effectively excluded from diseases having similar clinical features by highly sensitive and specific agglutination tests [4]. However, the presence of antibodies does not always indicate an active case of brucellosis, since humans from endemic areas often show weak serological responses [5].
In 2003 El-Sherbini et al. carried out a community-based survey in 2 villages in the Nile delta. The seroprevalence of brucellosis among humans was 1.7% and the diagnostic seropositivity level by STA was 1/320 [6]. The endemicity of brucellosis in our country necessitates isolation of the organism for confirmation of high STA positive titres. As for other fastidious pathogens, molecular detection by polymerase chain reaction (PCR) offers an alternative method of diagnosis of brucellosis. There are few studies concerning the use of PCR for diagnosis of brucellosis in humans [7–12].
We aimed to establish an accurate and sensitive PCR technique for the diagnosis of active human brucellosis for the first time at the Faculty of Medicine, University of Cairo and the National Research Centre, Cairo. PCR would provide rapid diagnosis of brucellosis, which is important for initiating specific treatment for patients. Furthermore, as brucellosis is a laboratory-acquired infection and may be associated with bioterrorism there are laboratory safety and wider public health implications to establishing such a test.
Methods
Patients
The study was conducted at the University of Cairo and the National Research Centre from July 2005 to September 2006. The sample was 50 patients from Imbaba Fever Hospital and the Tropical Disease Unit, University of Cairo Hospital. The limited number of patients included in the research is related to the budget provided. Consent was taken from patients; no refusal to participate was recorded. The case definition was: clinically suspected brucellosis [13], positive STA titre ≥ 1:160 and positive blood culture. For each patient we recorded demographic, occupational, clinical and risk factor data.
Laboratory tests
Blood samples were collected aseptically into 2 mL Na-citrate tubes and stored at –20 ºC until analysis.
Standard methods
The serological diagnosis was established by STA (Brucella antigen, Sanofi Diagnosis, Marnes La Coquette, France) using standard methods [3].
Blood cultures were processed with BACTEC 9050 aerobic blood culture bottles (Becton-Dickinson) and incubated for 21 days. Positive blood culture bottles were subcultured on Brucella blood agar with incubation in 5% CO2 at 37 ºC for up to 48 hours [2].
PCR methods
Two methods were used for extraction of DNA from clinical blood samples. The first method was a ready-made extraction kit (BioFlux, Fluxion Biosciences, San Francisco). The second by an extraction technique designed in-house by modification of the method described by Miller et al. [14], as follows.
- Blood was suspended in 0.5 mL of erythrocyte lysis solution (320 mM saccharose, 5 mM Mg2Cl, 1% triton X-100 and 10 mM tris HCl (pH 7.5), mixed, and centrifuged at 15 000 g for 2 min before discarding the supernatant. The leukocyte pellet was washed with 1 mL of sterile Milli-Q® water. For elimination of any remains of the haeme compound, 100 μL of an H2O2 (30% wt/wt) solution was added to the pellet and incubated for 2–5 min at room temperature. The H2O2 was removed with the tip of the pipette.
- Then 400 μL of nucleic lysis buffer (60 mM NH4Cl and 24 mM Na2 EDTA (pH 8.0) containing proteinase K (10 mg/mL) and sodium dodecyl sulfate (10%) was added to the pellet, mixed and incubated for 30 min at 55 ºC or overnight at 37 ºC with slow shaking. The samples were cooled at room temperature and 100 μL of ammonium acetate (7.5 M) was added to each tube. The tubes were shaken for 30 s, followed by centrifugation at 15 000 g for 10 min. The supernatant containing total DNA was transferred to a fresh tube. Two volumes of absolute alcohol were added and the tubes were inverted several times until the DNA was precipitated.
- DNA was recovered by centrifuging the samples at 15 000 g for 10 min, and the pellets were rinsed with 1 mL of 70% ethanol, dried and suspended in 40 μL of sterile Milli-Q® water. The concentration and purity of the DNA were then determined spectrophotometrically by readings of A260 and A280.
DNA amplification was done by 2 different PCR sets of primers: B4/B5 (223 bp) and JPF/JPR (193 bp). Most cases were carried out in duplicate. - Primers B4 (5'-TCGGTTGCCAATATCAA-3') and B5 (5'-CGCGCTTGCCTTTCAGGTCTG-3') were used to amplify a target sequence of 223 bp present on a gene encoding a 31 kDa B. abortus antigen. This sequence has been shown to be common to all Brucella biovars. For this PCR 50 μL of reaction mixture contained 10 mM tris HCl (pH 8.4), 50 mM KCl, 1 mM magnesium chloride, 200 μM each deoxyribonucleoside triphosphate (dATP, dGTP, dCTP, dTTP), oligonucleotides B4 and B5 (100 nM each), 1.25 μ of Taq polymerase (Boehringer), 2–4 μg of total DNA extracted from blood samples and 100 ng from the positive controls. The reaction was performed in a DNA thermocycler (model 2400, Perkin Elmer, Norwalk). After an initial denaturation at 93 ºC for 5 min, the PCR profile was set as follows: template denaturation at 90 ºC for 60 s, primer annealing at 60 ºC for 30 s and primer extension at 72 ºC for 60 s, for a total of 35 cycles, with a final extension at 72 ºC.CGCGCTTGCCTTTCAGGTCTG-3') were used to amplify a target sequence of 223 bp present on a gene encoding a 31 kDa B. abortus antigen. This sequence has been shown to be common to all Brucella biovars. For this PCR 50 μL of reaction mixture contained 10 mM tris HCl (pH 8.4), 50 mM KCl, 1 mM magnesium chloride, 200 μM each deoxyribonucleoside triphosphate (dATP, dGTP, dCTP, dTTP), oligonucleotides B4 and B5 (100 nM each), 1.25 μ of Taq polymerase (Boehringer), 2–4 μg of total DNA extracted from blood samples and 100 ng from the positive controls. The reaction was performed in a DNA thermocycler (model 2400, Perkin Elmer, Norwalk). After an initial denaturation at 93 ºC for 5 min, the PCR profile was set as follows: template denaturation at 90 ºC for 60 s, primer annealing at 60 ºC for 30 s and primer extension at 72 ºC for 60 s, for a total of 35 cycles, with a final extension at 72 ºC.
- Primers JPF (5'-GCGCTCAGGCTG-CCGACGCAA-3') and JPR (5'-ACC-AGCCATTGCGGTCGGTAA-3') were used to amplify a target sequence of 193 bp. Isolated DNA was amplified as described above, except for the concentrations of MgCl2 (3 mM) and primers (JPF/JPR, 300 nM each) [15]. This PCR consisted of an initial 4 min incubation step at 94 ºC, followed by 35 cycles with denaturation at 94 ºC, annealing at 60 ºC and extension at 72 ºC (each for 1 min) and a final 5 min extension step at 72 ºC.
For quality control the positive control was DNA extracted from Brucella colonies provided courtesy of the Naval American Medical Research Unit No. 3. The DNA was extracted from Brucella spp. colonies isolated from 2 different patients who suffered from brucellosis and whose laboratory results were positive by serology and blood culture. For the negative control all reagents were added except Brucella DNA.
The sample was considered positive when DNA with a molecular weight expected for the amplified product was observed after electrophoresis in 2% agarose with fluorescence in the presence of ethidium bromide (2 μg/mL). PCR amplification products were detected through visualization of bands under ultraviolet light.
Statistical analysis
All tabulated data were interpreted and analysed using SPSS, version 13. Independent t-test and chi-squared tests were used to detect significant differences (P < 0.05).
Results
Clinical data
The median age of the patients was 32 years (range 13–55 years); 36 (72%) were males. Of the 50 patients, 23 had a risk factor for brucellosis; 21 (42%) had contact with animals, either occupational [9 cases (18%) were farmers and butchers] or keeping animals near where they lived [12 (24%) cases]; 2 patients (4%) had a history of consuming unpasteurized milk. The exposure to risk factors was highly significant for brucellosis (P = 0.008).
Serology
Of the 50 patients, 6 (12%) had an STA titre of 1/160, 12 (24%) had 1/320, 16 (32%) had 1/640 and 16 (32%) had 1/1280 (Table 1).
Blood cultures
Brucella spp. were isolated in blood cultures from 15 patients who had not received antibiotic treatment at the time the specimen was collected. These patients were also positive by both PCR and serology with titres 1/1280.
PCR results
Using the ready-made extraction kits all samples gave negative results by the 2 protocols. Repetition of extraction using the in-house extraction technique was successful and PCR was positive in 35 samples (Figure 1). PCR was positive for all patients with positive blood cultures.
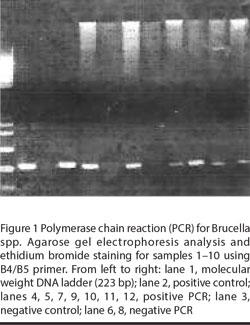
Evaluation of the technique
PCR positivity increased significantly with increasing seropositivity (P = 0.001). In cases with low STA positive titres 1/160, all PCR results were negative, out of 12 cases with titre 1/320, 7 cases were negative and out of 16 cases with titre 1/640, 2 cases were negative, while in the 16 cases with high titre 1/1280, all PCR results were positive (Table 1). Using positive blood culture as a gold standard [16], our PCR technique showed a sensitivity of 100%. Using STA titre > 1/160 as a gold standard [7] the sensitivity was 79% and the specificity was 100% (Table 2).
Discussion
In our study, the median age of patients was 32 years (range 13–55) and 72% were males. In a study that implemented laboratory-based surveillance in hospitals through Egypt, patients had a median age of 30 years (range 4–90) and 65% were males [1]. In Banha, Egypt, the age range was 23–56 years [16], while in Jordan the median age was 46 years (range 6–86) [5] and in Italy the median age was 42 years (range 16–47) [17]. We found that the risk factors for brucellosis were occupational exposure (farmers and butchers), drinking unpasteurized milk and history of contact with animals (P = 0.008). These data are consistent with Afifi et al.’s study on brucellosis in Egypt [1].
Although most researchers have used commercial kits for extraction of Brucella DNA [10,12] we failed to extract DNA using a commercial kit and this technique was discontinued. We used an in-house extraction method modified from the methods of Miller [14], which was successful and more economical. Lack of standardization of DNA extraction techniques and the difficulty in extraction from intracellular organisms might have made the use of the in-house extraction method preferable in our study. Diagnosis of human brucellosis in Egypt by PCR has not been done before.
Using blood culture as a gold standard, our PCR technique showed a sensitivity of 100%, in accordance with other authors [18,19]. This high sensitivity in culture-positive cases suggests that PCR may replace blood culture as the gold standard for acute brucellosis.
The sensitivity of PCR using titre > 1/160 as a gold standard was 79% and the specificity was 100%. Others who have tried to establish a PCR technique for diagnosis of human brucellosis found the sensitivity was 50% and the specificity 60% [10]. The existence of false negatives could be due to the presence of polymerase inhibitors. Many substances have been suggested as amplification inhibitors, including haemoglobin, urine, heparin, phenol and sodium dodecyl sulfate. In our study we used H2O2 to avoid this effect. Test sensitivity is also influenced by DNA extraction and detection procedures [10]. Other hypotheses to account for false-negative PCR results are the presence of a number of organisms below the detection limit and the degradation of target DNA in the samples [20].
The PCR positivity in our study increased significantly with increasing seropositivity. This can to some extent be explained by false positive serology results at titre 1/160 due to the endemicity of brucellosis in Egypt. This agrees with El-Sherbini et al. in Egypt who recommended increasing the seropositivity level of STA to 1/320 [6], while another study from Egypt used a positive agglutination titre > 1/160 as seropositive for brucellosis [16]. A study from Saudi Arabia showed that PCR was positive in 80% of symptomatic cases with titre 1/160 and the author recommended that titres > 1/160 should be reported as positive [21]. A study from Jordan also considered 1/160 as positive for serodiagnosis [5]. So we recommend performing a larger study in all governorates of Egypt to establish a titre for serodiagnosis of brucellosis in Egypt.
In conclusion, we have established an accurate and sensitive PCR technique for the first time in 2 important academic centres in Egypt: the Faculty of Medicine, University of Cairo and the National Research Centre. Our PCR technique proved to be both sensitive and specific. We recommend using PCR as an alternative to blood culture for diagnosis of acute brucellosis. Further studies are required to compare different techniques. We also recommend performance of a large-scale study to test this PCR technique for screening for brucellosis in Egypt.
References
- Afifi S et al. Hospital-based surveillance for acute febrile illness in Egypt: a focus on community-acquired bloodstream infections. American journal of tropical medicine and hygiene, 2005, 73(2):392–9.
- Chu MC, Weyant RS. Brucella. In: Murray PR et al., eds. Manual of clinical microbiology, 8th ed. Washington DC, American Society for Microbiology, 2003.
- Sirmatel F, Türker M, Bozkurt AI. Brusellozisin serolojik tanisinda kullanilan yontemlerin degerlendirilmesi [Evaluation of the methods used for the serologic diagnosis of brucellosis]. Mikrobiyoloji bulteni, 2002, 36:161–7.
- Mert A et al. The sensitivity and specificity of Brucella agglutination tests. Diagnostic microbiology and infectious disease, 2003, 46:241–3.
- Nimri LF. Diagnosis of recent and relapsed cases of human brucellosis by PCR assay. BMC infectious diseases, 2003, 3:5.
- El-Sherbini A et al. Seroprevalences and local variation of human and livestock brucellosis in two villages in Gharbia governorate, Egypt. Transactions of the Royal Society of Tropical Medicine and Hygiene, 2007, 101(9):923–8.
- Matar GM, Khneisser IA, Abdelnoor AM. Rapid laboratory confirmation of human brucellosis by PCR analysis of a target sequence on the 31-kilodalton Brucella antigen DNA. Journal of clinical microbiology, 1996, 34:477–8.
- Isabel MA et al. Rapid diagnosis of human brucellosis by peripheral-blood PCR assay. Journal of clinical microbiology, 1997, 35:2927–30.
- Morata P, Queipo-Ortuño MI, Colmenero JD. Strategy for optimizing DNA amplification in a peripheral blood PCR assay used for the diagnosis of human brucellosis. Journal of clinical microbiology, 1998, 36:2443–6.
- Navarro E et al. PCR assay for diagnosis of human brucellosis. Journal of clinical microbiology, 1999, 37(5):1654–5.
- Navarro E et al. Comparison of three different PCR methods for detection of Brucella spp. in human blood samples. FEMS immunology and medical microbiology, 2002, 34(2):147–51.
- Morata P et al. Development and evaluation of a PCR enzyme-linked immunosorbent assay for diagnosis of human brucellosis. Journal of clinical microbiology, 2003, 41:144–8.
- Surveillance of communicable diseases: a training manual. Alexandria, World Health Organization, Regional Office for the Eastern Mediterranean, 1998.
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic acids research, 1988, 16:1215.
- Leal-Klevezas DS et al. Single step PCR for detection of Brucella spp. from blood and milk of infected animals. Journal of clinical microbiology, 1995, 33:3087–90.
- Hafez AM et al. Rapid diagnosis of human brucellosis by peripheral blood PCR assay. Egyptian journal of medical science, 2003, 12(1):1–16.
- Iaria C et al. Live nativity and brucellosis, Sicily. Emerging infectious disease, 2006, 12(12):2001–2.
- Queipo-Ortuño MI et al. Rapid diagnosis of human brucellosis by peripheral blood PCR assay. Journal of clinical microbiology, 1997, 35:2927–30.
- Zerva L et al. Serum is the preferred clinical specimen for diagnosis of human brucellosis by PCR. Journal of clinical microbiology, 2001, 39:1661–4.
- Romero C et al. Evaluation of PCR and indirect enzyme-linked immunosorbent assay on milk samples for diagnosis of brucellosis in dairy cattle. Journal of clinical microbiology, 1995, 33:3198–200.
- El-Feki MG et al. Evaluation of culture, tube agglutination, and PCR methods for the diagnosis of brucellosis in humans. Medical science monitor, 2005, 11(11):MT69–74.








